At present, knowledge about the (eco-)evolutionary effects of polyploidy is largely based on either the comparison of polyploids with the contemporary descendants of their diploid ancestors and their synthetic tetraploids or on signatures of Whole Genome Duplications (WGDs) in present-day plant genomes. These comparative methods, although informative, suffer from potential biases introduced by evolution of both the polyploid and its ancestors after establishment. Experimental studies using artificial polyploids and their direct ancestors of lower ploidy are necessary to gain further insights into the mechanistic underlying of the short and long-term establishment and effects of polyploidy. We initiated several (eco-)evolutionary experiments wherein both conditions for initial establishment and long(er)-term evolution of polyploid genome characteristics can be studied.
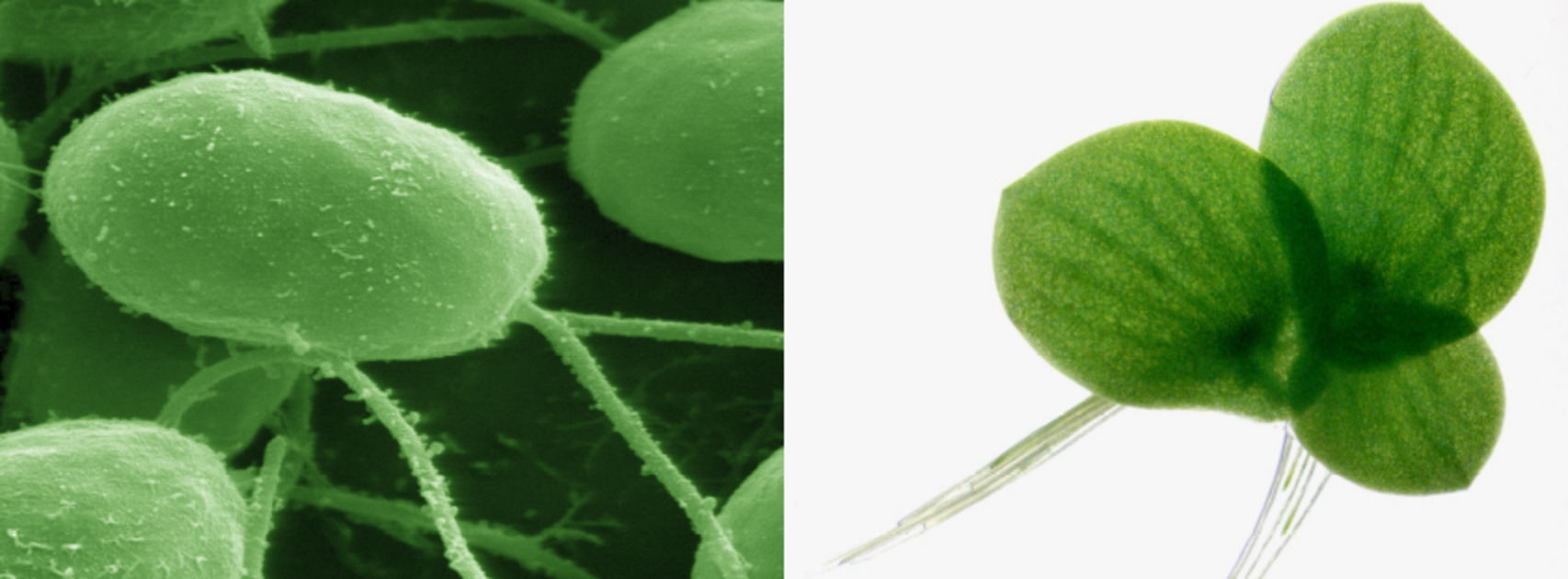
Up till now, only a few long(er)-term studies on polyploidisation have been conducted. These experimental studies used resynthesized yeast polyploids and have yielded some understanding into the short-term effects of polyploidisation on both adaptation and genome structure. However, no similar experiments have yet been conducted for the green lineage (green algae plus plants). Because polyploidy seems to be especially preponderant in the green lineage, we use the unicellular green alga Chlamydomonas reinhardtii and the small aquatic macrophyte Spirodela polyrhiza to study polyploidization in the green lineage. Both organisms are characterised by a high vegetative growth rate, relatively small genome sizes, limited space requirements, easy culture establishment in the laboratory, and a rich history as a model system, making them well suited to study the functional effects of WGD and long term evolution.
We use Chlamydomonas to study (1) differences in adaptation rate between diploid and haploid lines and (2) the allo(poly)ploid plasticity hypothesis. This hypothesis states that allopolyploid species might utilize the expression patterns of both parents. Spirodela is used to investigate (1) the immediate phenotypic effect of WGD in genetically diverse duckweed strains, (2) ploidy induced changes in mutation rate and spectrum using mutation accumulation, (3) differences in the rate of adaptation under environmental change (evolve and resequence experiments) and (4) population dynamics and establishment of tetraploids in a diploid population.
Ultimately, we aim to obtain insight in the adaptive potential of polyploids in changing environments. To achieve this, we complement this in vivo experimental approach with in silico experiments based on so-called digital organisms (DOs) running on artificial genomes. The obtained results of both approaches will be cross-referenced with each other.