Early 2021, seven Ghent University research groups received funding so that they can develop projects throughout the next 7 years. Our research group was one of them. The Methusalem program offers the most prestigious and extensive funding from the Ghent University Special Research Fund. A total of 27.5 million euros have been allocated for a period of 7 years. With this funding, the laureates and their research groups can further strengthen their international benchmark position and ensure that Ghent University improves its already significant reputation in their respective fields.
Summary of the Methusalem proposal: Polyploidy, i.e., the possession of multiple sets of chromosomes as a consequence of whole-genome duplication (WGD), has been known for a long time, especially in plants. Although polyploidy is rarer in animals, there are also numerous cases of polyploid insects, fishes, amphibians, and reptiles. For a long time, ancient polyploidy, dating back millions of years, was much less well documented and it was not until the advent of genomics and whole genome sequencing that it became clear that the significance of polyploidy extends across all eukaryotes, and even prokaryotes, from ancient history to the recent past. Most, if not all, extant species (including our own) carry the signature of at least one ancient WGD. Because of their often-enhanced phenotypic appearance, polyploidy has also been a key force in the origin and success of most crops. Artificial polyploidization of crops can increase yield, consumer satisfaction, and specific nutrients, thereby improving food security, a critical goal given the Earth’s expanding population and limited arable land. In addition to occurring in whole organisms, programmed or unprogrammed events can increase the ploidy of specific somatic cells and cell lineages. In humans, for instance, polyploid cells keep the human heart beating, and are essential for repair of the most regenerative organ in the human body; the liver. Polyploid cells are important in the development of structures including trichomes, fruits, and root nodules. Acute, induced polyploidy in individual cells or tissues can also occur in response to tissue stress and in disease. Finally, unprogrammed ploidy increases are now known to be among the most common events in human tumor growth. In conclusion, polyploidy is a driving force in organismal and sub-organismal evolution and elucidating the consequences of WGD at multiple levels is key to understanding global patterns of biodiversity and ecology, as well as cellular fates, physiology, and metabolism. Although the implications of polyploidy range from cells to ecosystems and from agriculture to human health, polyploidy remains understudied in many contexts, and its roles and impact in biological processes and across phylogeny are unclear.
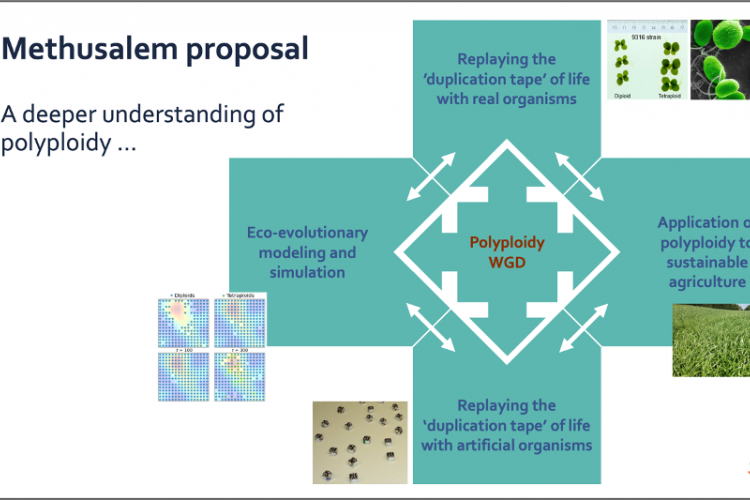
One recurring theme is the link between polyploidy and stress. It is known that stress can trigger polyploidy, but there are also strong indications that polyploidy confers a selective advantage under stressful conditions, such as during environmental turmoil. In this research project, we want to build on our current expertise in polyploidy and WGD by unraveling the mechanistic complexities underlying polyploidy under stressful conditions. To this end, we want to use a holistic interdisciplinary approach integrating genomics, experimental evolution, modeling, and Artificial Intelligence (AI).
One major line of research will be experimental evolution, where we will replay ‘the genome duplication tape of life’ in both biological and artificial systems. For two different biological model systems, namely the green alga Chlamydomonas reinhardtii and the fast-growing angiosperm Spirodela polyrhiza (greater duckweed), we will initiate and run long-term evolutionary (and resequencing) experiments, where we will compare polyploid populations with non-polyploid populations and see how they adapt to different/stressful environments. Furthermore, we will complement these experiments with ‘in silico’ simulations based on so-called digital organisms running on (duplicated) artificial genomes encoding gene regulatory networks. Complementary modelling approaches will also be employed to study the effects of polyploidy from an eco-evolutionary dynamic perspective, integrating population and quantitative genetics with ecological interactions. By evaluating and integrating the results obtained from these different in vivo and in silico experiments, we will obtain important novel insights in the adaptive potential of polyploids under stressful conditions or during times of environmental change.
A second major line of research entails the application of polyploidy to crop domestication. Here, we want to gain deeper insight into the mechanisms underlying genome dominance in interspecific plant hybrids at different ploidy levels using genomics and transcriptomics. Understanding rules underlying increases in ploidy and genetic diversity may help breeders to choose breeding materials on predicted compatibility of biological networks underlying complementary growth types, necessary fertility characteristics and responses to environmental stress. In the current project, we will be studying the first effects of polyploidy in the Festuca-Lolium complex. Polyploid ryegrass is the basis of common tetraploid ryegrass breeding programs because of its better digestibility. Festuca species on the other hand are more drought resistant. Combining the beneficial traits of digestibility for cattle in Lolium and drought resistance in Festuca through interspecific hybridization between tetraploid genotypes of Lolium and tetraploid genotypes of Festuca results in higher annual dry matter yield and good digestibility. However, the Festuca-Lolium complex displays intra- and interspecies differentiation in growth habit and survival strategies. Furthermore, a disadvantage of intergeneric hybrids is a reduced seed set. Aberrations in chromosome numbers, and competition of parental or sub-genomes is common, and several studies have shown that in early generations after interspecific hybridisation, chromosomes of Festuca are gradually replaced by that of Lolium. The mechanisms underlying this genome dominance of one parental genome over the other are unknown. For future successful breeding of crops with better traits but keeping high seed yields, we need a better understanding about genome competition and genome dominance in hybrid species. Studying an extensive breeding population of the Festuca-Lolium complex through genome sequencing, transcriptomics, and epigenetics should provide further insight into the consequences of polyploidy in (crop) species.
Collaborators
- Prof. Olivier De Clerck, from the Phycology lab, Department of Biology, Faculty of Science, Ghent University, Ghent, Belgium Link here
- Prof. Dries Bonte, from the Spatial Ecology and Evolution group, Department of Biology, Faculty of Science, Ghent University Link here
- Profs. Pieter Simoens and Pieter Audenaert, from the Internet Technology and Data Science Lab, Faculty of Engineering and Architecture, Ghent University Link here
- Drs. Isabel Roldan, Leen Leus, and Tom Ruttink, from the Research Institute for Agriculture, Fisheries, and Food (ILVO), Ghent, Belgium Link here